Generative AI in Medical Images
Generative AI is a field with the potential to create new nonexistent/synthetic content that is eye-catching and eye-deceiving at the same time. This evolving part of AI has touched many domains, including text, audio, images, videos, etc. Even a combination of these domain types can be synthetically stitched together, which is very difficult to catch by the human eye. Amazingly, this mind-blowing creative content can be generated using the knowledge acquired from the existing data patterns. In other words, generative AI models are capable of using just the raw data to create new content with overall similar but not identical, rather unique information. This research area has experienced many breakthroughs in the past years and is currently the most trending topic in tech talks. Starting from 3D animations, gaming, media, and advertising, it has reached many more critical and crucial applications, such as healthcare.
Generative AI has the potential to transform and complement clinicians' abilities and hence, can improve patient care, drug discovery, and diagnostics. In addition, it can assist in overcoming the challenges faced by medical image analysis researchers due to lack of data, especially for some rare to find abnormalities. Including generative AI in clinical treatments provides various benefits, such as increased training samples for undersampled medical image classes, exemption from the procedures involved in collecting sensitive data and related privacy concerns, and personalized plans for retrospective treatments based on the patient's medical history. Apart from new content generation, the image-to-image translations supported by generative models introduce new ways to explore some significant tasks, such as noise/artifacts removal and domain adaptation. This article discusses a similar artifacts removal technique using CycleGAN which explores the abilities of generative models in transforming the uninformative colonoscopy image frames into clinically significant frames.
This discussion is based on “Can Adversarial Networks Make Uninformative Colonoscopy Video Frames Clinically Informative? (Student Abstract)”, published in AAAI 2023. The article covers the concept, implementation details, and code execution guidelines.
Brief Introduction
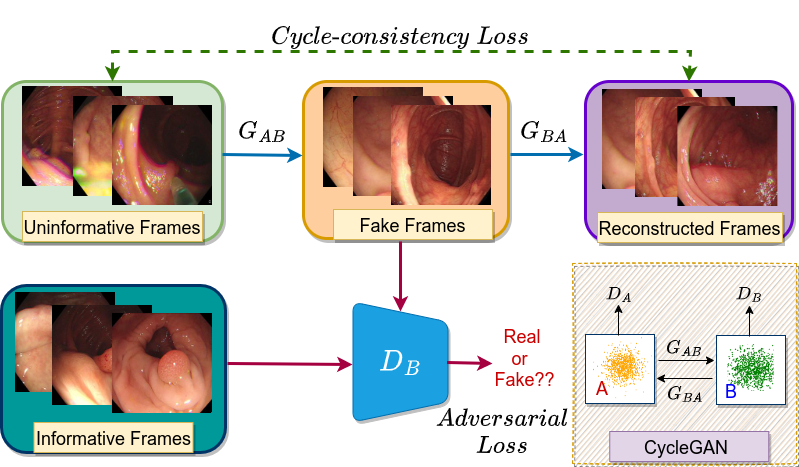
Colonoscopy videos are acquired using a coloscope mounted with a camera. During the procedure, a large amount of video frames are captured. However, not all frames satisfy the image quality requirements necessary for correct pathological diagnosis. The factors affecting the image quality include improper patient preparation and abrupt camera movements that introduce unwanted artifacts such as ghost colors, interlacing, and motion blur. The presence of these artifacts can hinder both manual and automated diagnosis of serious abnormalities like colorectal cancer. To address the issue of insignificant/uninformative frames and extract obscured significant/informative details, an adversarial network based approach is discussed, inspired by the unpaired image-to-image translation supported by CycleGAN. In medical imaging, obtaining paired sets of data with uninformative and informative counterparts is difficult. Hence, it is important to adopt an unpaired approach that can learn from the data distribution of one domain (a pool of significant frames) and translate the images to another domain such that they are indistinguishable from the former. In short, we have a domain A containing uninformative frames and a domain B with informative frames. We aim to translate domain A frames into domain B frames such that the translated data is indistinguishable from the original insignificant frames.
Domain A: Uninformative colonoscopy frames
Domain B: Informative colonoscopy frames
Aim: Translate Domain A to Domain B
Note that we do not have paired data, i.e., no mapping between the images of two domains. This major challenge which we often come across in medical images, is gracefully handled by the concept introduced in CycleGAN. Like a general GAN, it has a generator and a discriminator but notice their count difference. It has two generators and two discriminators. Let’s discuss their specific tasks. The generator \(G_{AB}\) learns the mapping function that targets to translate domain A frames to domain B frames, whereas the generator \(G_{BA}\) learns to translate domain B frames to domain A frames. Both generators have their corresponding discriminator that aims to distinguish the synthetic images from the real ones.
\[G_{AB}: A \rightarrow B\] \[G_{BA}: B \rightarrow A\]\(D_B:\) Distinguishes B from \(G_{AB}(A)\)
\(D_A\): Distinguishes B from \(G_{BA}(B)\)
Objective Function
The objective function of CycleGAN comprises two components: *Adversarial loss* and *Cycle-consistency loss*. The concept behind adversarial loss is the same as in other GAN-based approaches. The generators try to *fool* their corresponding discriminators so that they are not able to distinguish between the synthetic images with the real ones. The adversarial loss helps calculate the distance between the data distributions related to the generated and the original frames. It can be defined as:
\(L_{adv}(G_{AB}, D_B) = \mathbb{E}_{b\sim p_{data}(b)}[(D_B (b)-1)^2] + \mathbb{E}_{a\sim p_{data}(a)}[(D_B(G_{AB}(a)))^2]\)
An important component of the CycleGAN is the cycle-consistency loss. Due to unmapped data in both domains, the same set of images can be mapped randomly to any possible permutations in the other domain. To avoid this situation, cycle-consistency loss plays an important role. It can be defined as:
\[L_{cyc}(G_{AB}, G_{BA}) = \mathbb{E}_{a\sim p_{data}(a)}[\lVert G_{BA}(G_{AB}(a))-a\rVert_1] + \mathbb{E}_{b\sim p_{data}(b)}[\lVert G_{AB}(G_{BA}(b))-b\rVert_1]\]The idea is:
If we translate the domain A (source) distribution to the domain B (target) distribution and then again try to translate the domain B (target) distribution to the domain A (source) distribution, we should be able to obtain samples from domain A (source).
Implementation Details
The PyTorch implementation of CycleGAN is available at https://github.com/junyanz/pytorch-CycleGAN-and-pix2pix. The dataset used in this work (SUN Database) can be obtained on sending a request as per the instructions mentioned on http://sundatabase.org/. Also, to obtain the annotations for informative and uninformative frames, you can refer to this paper and can contact me at this mail id. Once you download the code and the dataset, create a folder ./data inside your downloaded code’s dataset folder. Then create subfolders testA, testB, trainA, and trainB. Place your uninformative frames pertaining to the train and test set in the trainA and testA subfolders, respectively. Do the same for informative frames and place them in the trainB and testB folders. To run the files using the docker image, you can use the image available at vanshalisharma/my_kubernetes_image:cgan.
Let’s train the CycleGAN using the below command:
python3 train.py --dataroot ./datasets/data --name colonframes --model cycle_gan --display_id 0
Here, you can give any name for --name option for your model. By default, the generators follow 9 ResNet blocks architecture, and the discriminators use PatchGAN-based architecture. You can explore different architectures using --netG and --netD argument options, and this argument list can be found in *options/base_options.py*. One example of usage is given below:
python3 train.py --dataroot ./datasets/data --name colonframes --model cycle_gan --display_id 0 --netG unet_256
To test the model, execute the following command:
python3 test.py --dataroot ./datasets/data/testA --name colonframes --model test --no_dropout --num_test 1000
Remember to replace –num_test option with the count of images you want to test.
Strengths and Limitations
Now the most important question is, “How good are the generated images?” (This question is critical in medical image analysis!). In the paper “Can Adversarial Networks Make Uninformative Colonoscopy Video Frames Clinically Informative? (Student Abstract)”, YOLOv5 has been adopted to verify the quality of images obtained from the generator. It has been observed that the YOLOv5 performance got enhanced when translated informative frames were used. The artifacts, such as ghost colors, low-illumination, and fecal depositions, are satisfactorily handled using CycleGAN. However, motion blur and interlacing are still present in the translated images.
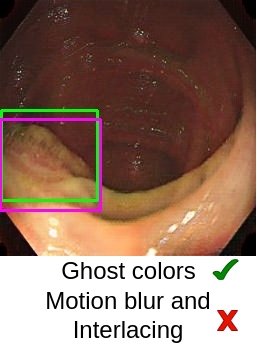
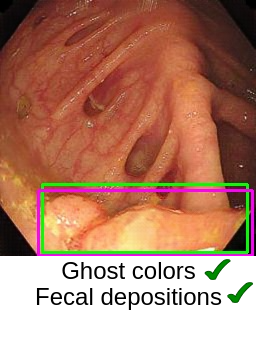
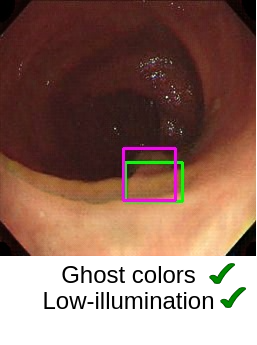
In medical images, lesion-characterizing features are significantly important as these features decide the pathological (such as cancerous/non-cancerous) condition. Therefore, the generated images should retain such features. For example, some of the key features observed during analysis of colon polyps are their shape, color, and texture. Although generative AI has shown promising results but it requires more research and analysis in the medical domain to rely on such systems on a large scale.